
VIRALERT project
Environmental-based epidemiology for preparedness and early detection of viral epidemics
Background
Recently, viral epidemics have increased in both size and frequency. Global inter-connectivity has undeniably facilitated the spreading of infectious diseases across continents. Climate change is also affecting the geographic distribution of disease vectors and the interface between humans and animals, thus providing more opportunities for pathogens to jump between species.
Viral infections such as SARS-CoV-2 cause a wide array of symptoms, and severe cases are often just the tip of the iceberg. This may result in a lag between the emergence of pathogens and their detection by classical surveillance systems that are reliant on clinical signs.
While the emergence and spread of SARS-CoV-2 illustrated the current challenges in infectious disease surveillance, management and control, it also promoted innovations. Although wastewater surveillance had already been used in the context of poliovirus elimination strategies, the COVID-19 pandemic offered the opportunity to further highlight its advantage in providing a cost-effective and non-invasive picture of pathogen circulation in an entire population.
Nevertheless, from the point of view of a One Health context that recognises the interdependency of human, animal and environment health, surveillance needs to evolve even further. Wastewater mainly captures the human microbiome but disregards animal populations. Surface water epidemiology, exploiting environmental faecal contamination as a marker of wild and domestic animal health, has been seldom explored to bridge this gap.
Aims
Building on the long-standing expertise of the Clinical and Applied Virology group at the LIH in human and animal disease surveillance, and on previous collaborations between the LIH and LIST in the domain of infectious disease surveillance in the environment (cfr FNR funded projects “SENSORLUX” and “CORONASTEP+”), the VIRALERT project proposes an innovative integrative approach for monitoring the circulation of viruses with high epidemic and pandemic potential in a non-invasive cost effective manner.
To this end, VIRALERT aims to
- monitor an extended range of viruses in wastewater, including enteric and respiratory viruses,
- monitor population health in a post-pandemic context in which the circulation of other viruses has been affected by pandemic containment measures,
- cover wild and domestic animal health by investigating viral populations in freshwater,
- explore the potential of various technologies with differing sensitivity and specificity characteristics for the molecular detection and sequencing of influenza viruses, human and bovine respiratory syncytial viruses, seasonal human and animal coronaviruses, enteroviruses, noroviruses and mosquito-borne flaviviruses and
- take advantage of next generation sequencing (NGS) technologies to explore the possibility to detect a wider range of viruses.
In the long run, enhanced environmental surveillance will provide an additional tool for public and animal health authorities that complements existing surveillance systems and contributes to decreasing the burden of infectious diseases.
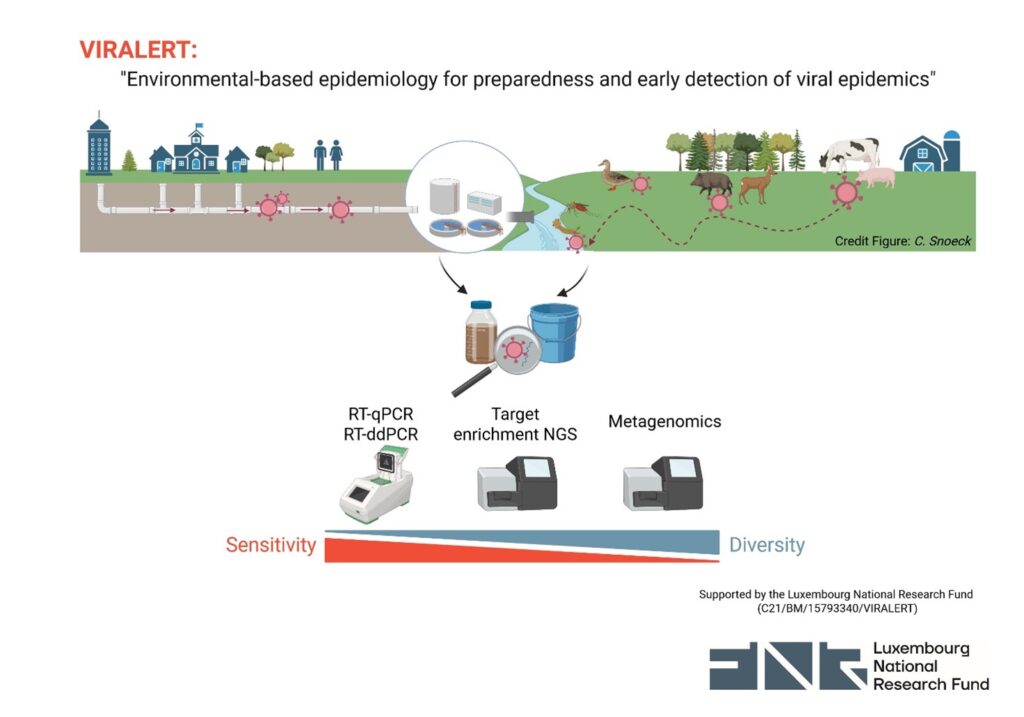
Graphical view of the VIRALERT project overarching scheme and technologies employed (created with BioRender).
Abbreviations: RT-qPCR real-time reverse transcription polymerase chain reaction; RT-ddPCR reverse transcription digital droplet polymerase chain reaction; NGS next generation sequencing
CONTACT
Hübschen
Partners



Funding

Supported by the Luxembourg National Research Fund (C21/BM/15793340/VIRALERT)
