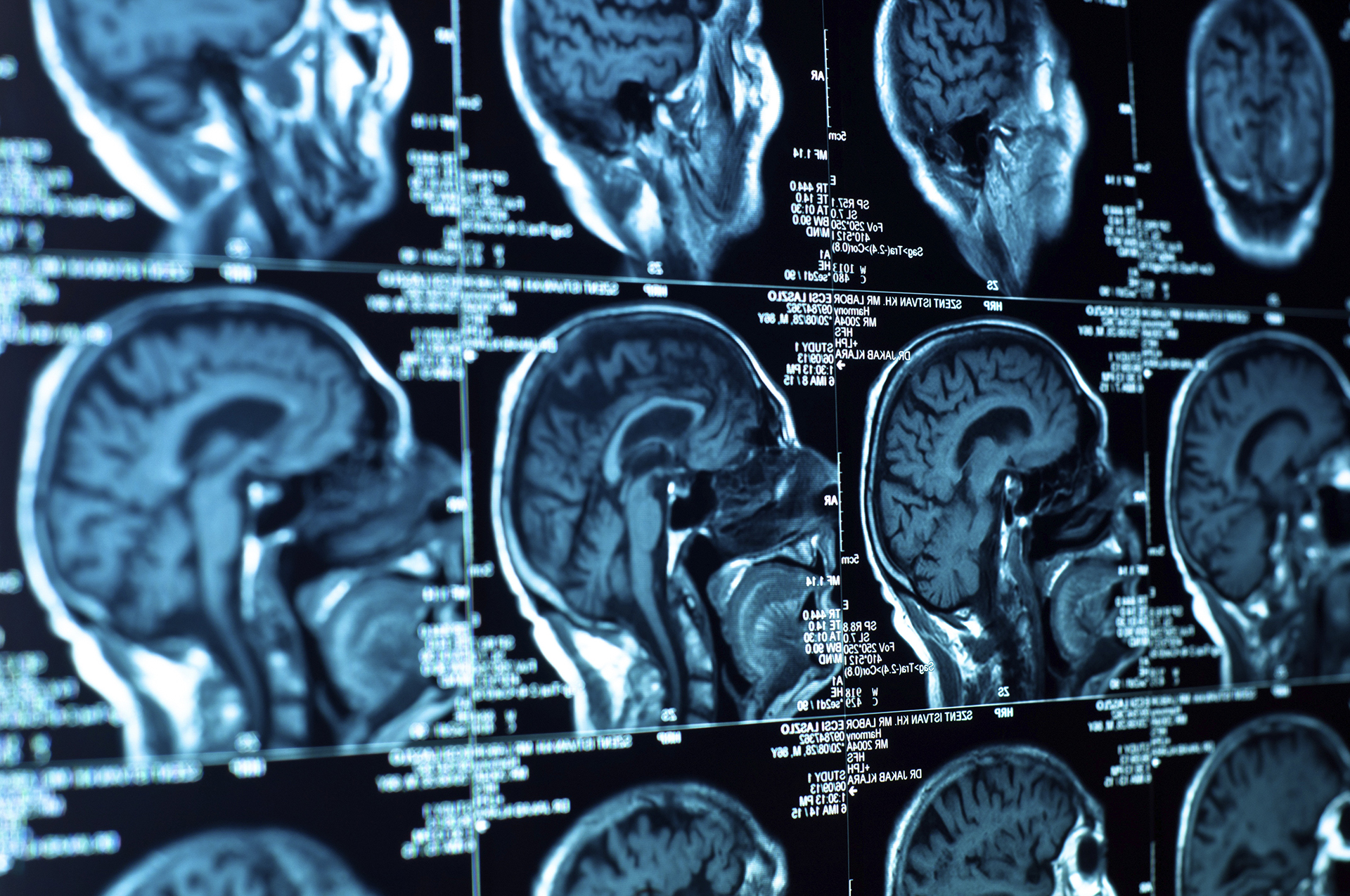
Advancing personalised cancer treatment for patients with brain tumours. Help change the future of Brain Cancer treatment!
Click on the projects for more information.
August 2021 – August 2024
Project PI: Michel Mittelbronn, MD, PHD
National Center of Pathology (NCP) & Luxembourg Center of Neuropathology (LCNP)
Laboratoire National de Santé
Abstract:
Radiographic imaging in glioma is routinely recorded and provides a non-invasive clinical measure before tumor resection. It is further used to monitor patients throughout their treatment and follow-up and can help to identify early relapse risk. Current standard-of-care (SOC) is neurosurgical resection, as radically as possible, radiotherapy (RT) and oral chemotherapy using temozolomide (TMZ). Guidelines for therapy focus on first-line therapy only. However, timing of second-line interventions has very important implications: Waiting too long can potentially result in tumor growth into brain areas making additional neurosurgical intervention necessary. Intervening too early, or treating pseudo-progression, exposes patients to morbidity risk and reduces quality of life.
The Mi*EDGE project is based on existing mathematical models of glioma invasion that predict tumor cell behavior based on critical density (“allee-effect”) and local metabolic-driven phenotypic plasticity (“go or grow” decision). It is aiming at predicting whether malignant cells at the margins of surgical resection transform into diffusely invasive or tumor-forming proliferative phenotypes by integrating clinical imaging and molecular pathology data, e.g. DNA methylation, multiplex-histology.
Thus, the initial investigations of the Mi*Edge consortium will be based on analysing the imaging time-points prior to operation, follow-up examinations after operation throughout the course of the disease and will include monitoring of disease progression involving MR images after re-surgery, re-RT or re-challenge with chemotherapy. We will test whether pre-surgical neuroimaging, when included into those models, can predict biological behavior of cells at the tumor edges, and if imaging after primary therapy will reflect the prediction with either diffuse tumor invasion, local relapse, or reactive changes (pseudo-progression). A number of specialized MRI scan components, such as perfusion weighted imaging (DSC-MRI), diffusion weighted or tensor imaging (DWI/ DTI) and specific contrast enhancement (DCE) will further help Mi*Edge to differentiate between unequivocal tumor progression and pseudoprogression.
Patient selection criteria and data collected:
Patients diagnosed with gliomas. The MRI scan types collected: i) Baseline MRI parameters, e.g. pre-/post-contrast T1-weighted, T2-weighted and T2-FLAIR and ii) Advanced MRI sequences, e.g. DSC-MRI, DWI/ DTI, DCE-MRI. MRI scans are anonymized by deidentification of textual data and brain extraction in the acquired cranial scans, preventing facial reconstruction/recognition.
April 2021 – ongoing
Project PIs: Simone Niclou, PHD, Anna Golebiewska, PHD & Olivier Keunen, PHD
NORLUX Neuro-Oncology laboratory and Translational Radiomics group
Department of Cancer Research
Luxembourg Institute of Health (LIH)
Abstract:
Radiographic imaging in glioma is routinely acquired, often providing the first non-invasive clinical impression (prior to biopsy or excision) and offering continuous monitoring throughout treatment and follow-up. The initial goals of iGLASS are based on the imaging timepoints, i.e., baseline pre-operative and follow-up examinations performed throughout the course of disease. At baseline, iGLASS will investigate i) distinct spatial tumor distributions of molecular characteristics, ii) mutational burden, and iii) tumor propensity to hypermutation following DNA-alkylating chemotherapy. Follow-up imaging studies linked to clinical annotation or recurrent tissue samples with molecular profiling will support investigation of i) radiogenomic biomarkers downstream of treatment, ii) longitudinal molecular alterations, and iii) treatment-induced hypermutation changes associated with worse outcome. To achieve this, iGLASS focuses on available imaging data corresponding to existing clinical and molecular data of the GLASS consortium contributors (https://glass-consortium.org/).
Patient selection criteria and data collected:
Patients diagnosed with gliomas that underwent two or more consecutive operations and were included in the GLASS consortium cohort for molecular analysis. The imaging data (MRI, CT, PET scans) will be collected and pseudonymised. The MRI scan types collected: i) Baseline MRI parameters, e.g. pre-/post-contrast T1-weighted, T2-weighted and T2-FLAIR and ii) Advanced MRI sequences, e.g. DSC-MRI, DWI/ DTI, DCE-MRI. MRI scans are anonymized by deidentification of textual data and brain extraction in the acquired cranial scans, preventing facial reconstruction/recognition.
Project website: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7566469/
April 2021 – December 2025
Project PIs: Olivier Keunen, PHD & Simone Niclou, PHD
NORLUX Neuro-Oncology laboratory and Translational Radiomics group
Department of Cancer Research
Luxembourg Institute of Health (LIH)
Abstract:
Radiographic imaging in glioma is routinely acquired throughout the management of this deadly disease. Proper detection and delineation of the tumor are important to establish diagnostic, assess the extent of the disease, plan therapy, notably radiotherapy, and assess the effect and efficacy of treatments.
The Federated Tumor Segmentation project addresses the challenging task of automatically delineating glioma, at the time of diagnosis and throughout the treatment. The task is rendered difficult by the heterogeneity of tumors that come in a variety of shapes and compositions. Artificial intelligence can help in this regard, but for the models to learn enough to reliably delineate tumor sub-regions, they need to see a large number of cases. Collecting large cohorts of patient imaging data is however difficult because of associated confidentiality and legal restrictions. So, to address this challenge, the project uses an innovative concept called Federated Learning, through which efficient models can be trained without the need to share data. With this approach, a basic model is first trained centrally before it is distributed to all participating hospitals sites. The remote sites assess the performance of the model on their local data and send the aggregator the numerical data needed to improve the model iteratively, but without ever sharing the original imaging data.
Patient selection criteria and data collected:
Patients diagnosed with gliomas for which pre-operative and follow-up MRI scans have been collected. The MRI scan types collected are baseline and follow-up scans, including pre-/post-contrast T1-weighted, T2-weighted and T2-FLAIR. Scans are anonymized by de-identification of textual data and brain extraction in the acquired cranial scans, preventing facial reconstruction/recognition.
Project website: https://www.nature.com/articles/s41467-022-33407-5
Feb 2022 – Dec 2024
Project PI: Alessandro Michelucci, PHD
Neuro-Immunology Group
Department of Cancer Research
Luxembourg Institute of Health (LIH)
Abstract:
Glioblastoma is an incurable malignant brain tumour that urgently needs new diagnostic and therapeutic approaches. A key challenge for clinical management of Glioblastoma is its highly heterogeneous nature, with heterogeneity between patients and within a single tumour representing important barriers for current therapies. The interactions between neoplastic cells and their microenvironment, including immune cells, affect Glioblastoma growth, patient survival, and response to therapy. Among non-neoplastic cells, immune cells called tumour-associated microglia/macrophages, which are mainly either resident parenchymal microglia or peripheral-monocyte derived cells, represent the majority of cells in the tumour microenvironment. Monocytes recruited from the bloodstream to the tumour site are associated with a worse prognosis compared to brain tumours with low peripheral immune cell infiltration. As tumours progress to malignancy, mediators produced by neoplastic cells and the tumour microenvironment are released into the bloodstream. These factors induce the mobilization, education and recruitment of peripheral monocytes to the tumour site, which differentiate into macrophages and stimulate angiogenesis, enhance tumour cell migration and suppress anti-tumour immunity. Further, these mediators induce severe systemic immunosuppression where, for example, peripheral lymphocytes display prominent dysfunctions and myelopoiesis is redirected from its normal pathway. Considering the tight crosstalk between the tumour in the brain and the immune components in the circulation, we here aim to analyse the blood immune cellular constituents of Glioblastoma patients, with a special focus on the monocytic populations. Taken together, the immunological profile of the blood in patients with Glioblastoma represents a signature of the tumour, which may serve for diagnosis, patient classification, prediction of immunotherapy efficacy and ultimately the discovery of novel targets.
Patient selection criteria and data collected:
Patients diagnosed with glioblastomas. The use of immune cells isolated from patients’ blood for molecular analysis.
Project website: https://www.lih.lu/en/research-scope/research-department/department-of-cancer-research/neuro-immunology-group/
| Cookie | Duration | Description |
|---|---|---|
| _GRECAPTCHA | 5 months 27 days | This cookie is set by the Google recaptcha service to identify bots to protect the website against malicious spam attacks. |
| cookielawinfo-checkbox-advertisement | 1 year | Set by the GDPR Cookie Consent plugin, this cookie is used to record the user consent for the cookies in the "Advertisement" category . |
| cookielawinfo-checkbox-analytics | 1 year | Set by the GDPR Cookie Consent plugin, this cookie is used to record the user consent for the cookies in the "Analytics" category . |
| cookielawinfo-checkbox-functional | 1 year | The cookie is set by the GDPR Cookie Consent plugin to record the user consent for the cookies in the category "Functional". |
| cookielawinfo-checkbox-necessary | 1 year | Set by the GDPR Cookie Consent plugin, this cookie is used to record the user consent for the cookies in the "Necessary" category . |
| cookielawinfo-checkbox-others | 1 year | Set by the GDPR Cookie Consent plugin, this cookie is used to store the user consent for cookies in the category "Others". |
| cookielawinfo-checkbox-performance | 1 year | Set by the GDPR Cookie Consent plugin, this cookie is used to store the user consent for cookies in the category "Performance". |
| CookieLawInfoConsent | 1 year | Records the default button state of the corresponding category & the status of CCPA. It works only in coordination with the primary cookie. |
| JSESSIONID | session | The JSESSIONID cookie is used by New Relic to store a session identifier so that New Relic can monitor session counts for an application. |
| PHPSESSID | session | This cookie is native to PHP applications. The cookie is used to store and identify a users' unique session ID for the purpose of managing user session on the website. The cookie is a session cookies and is deleted when all the browser windows are closed. |
| viewed_cookie_policy | 1 year | The cookie is set by the GDPR Cookie Consent plugin to store whether or not the user has consented to the use of cookies. It does not store any personal data. |
| Cookie | Duration | Description |
|---|---|---|
| bcookie | 1 year | LinkedIn sets this cookie from LinkedIn share buttons and ad tags to recognize browser ID. |
| bscookie | 1 year | LinkedIn sets this cookie to store performed actions on the website. |
| lang | session | LinkedIn sets this cookie to remember a user's language setting. |
| li_gc | 5 months 27 days | Linkedin set this cookie for storing visitor's consent regarding using cookies for non-essential purposes. |
| lidc | 1 day | LinkedIn sets the lidc cookie to facilitate data center selection. |
| pll_language | 1 year | The pll _language cookie is used by Polylang to remember the language selected by the user when returning to the website, and also to get the language information when not available in another way. |
| UserMatchHistory | 1 month | LinkedIn sets this cookie for LinkedIn Ads ID syncing. |
| vOTSXIlGdxFUt | 1 day | No description |
| xwNvEASRYf_ | 1 day | No description |
| Cookie | Duration | Description |
|---|---|---|
| _ga | 2 years | The _ga cookie, installed by Google Analytics, calculates visitor, session and campaign data and also keeps track of site usage for the site's analytics report. The cookie stores information anonymously and assigns a randomly generated number to recognize unique visitors. |
| _ga_ZM94YP9CD3 | 2 years | This cookie is installed by Google Analytics. |
| _gat_gtag_UA_16961320_1 | 1 minute | Set by Google to distinguish users. |
| _gid | 1 day | Installed by Google Analytics, _gid cookie stores information on how visitors use a website, while also creating an analytics report of the website's performance. Some of the data that are collected include the number of visitors, their source, and the pages they visit anonymously. |
| AnalyticsSyncHistory | 1 month | Linkedin set this cookie to store information about the time a sync took place with the lms_analytics cookie. |
| CONSENT | 2 years | YouTube sets this cookie via embedded youtube-videos and registers anonymous statistical data. |
| Cookie | Duration | Description |
|---|---|---|
| _fbp | 3 months | This cookie is set by Facebook to display advertisements when either on Facebook or on a digital platform powered by Facebook advertising, after visiting the website. |
| fr | 3 months | Facebook sets this cookie to show relevant advertisements to users by tracking user behaviour across the web, on sites that have Facebook pixel or Facebook social plugin. |
| IDE | 1 year 24 days | Google DoubleClick IDE cookies are used to store information about how the user uses the website to present them with relevant ads and according to the user profile. |
| test_cookie | 15 minutes | The test_cookie is set by doubleclick.net and is used to determine if the user's browser supports cookies. |
| VISITOR_INFO1_LIVE | 5 months 27 days | A cookie set by YouTube to measure bandwidth that determines whether the user gets the new or old player interface. |
| YSC | session | YSC cookie is set by Youtube and is used to track the views of embedded videos on Youtube pages. |
| yt-remote-connected-devices | never | YouTube sets this cookie to store the video preferences of the user using embedded YouTube video. |
| yt-remote-device-id | never | YouTube sets this cookie to store the video preferences of the user using embedded YouTube video. |
| yt.innertube::nextId | never | This cookie, set by YouTube, registers a unique ID to store data on what videos from YouTube the user has seen. |
| yt.innertube::requests | never | This cookie, set by YouTube, registers a unique ID to store data on what videos from YouTube the user has seen. |
| Cookie | Duration | Description |
|---|---|---|
| DEVICE_INFO | 5 months 27 days | No description |
| ln_or | 1 day | No description |
